Navigating STREAMLINE Output
This section covers the different outputs generated by STREAMLINE. The sections below will use the demo run of STREAMLINE on two demonstration datasets as a specific example for navigating the output files generated.
Notebooks
During or after the notebook runs, users can inspect the individual code and text (i.e. markdown) cells of the notebook. Individual cells can be collapsed or expanded by clicking on the small arrowhead on the left side of each cell. The first set of cells include basic notebook instructions and then specify all run parameters (which a user can edit direclty within the notebook). Later cells run the underlying STREAMLINE code for up to 9 phases, plus output folder cleaning (and downloading output files in the case of the Colab Notebook).
These later code cells will automatically display many of the notifications, results, and output figures generated by STREAMLINE.
As mentioned, the Google Colab notebook will automatically download the output folder as a zipped folder, as well as automatically open the testing and replication PDF reports on your computer. Users can then extract this downloaded output folder and view all individual output files arranged into analysis subdirectories. You can also view output files in Google Colab by opening the file-explorer pane on the left side of the notebook.
Experiment Folder (Hierarchy)
After running STREAMLINE you will find the ‘experiment folder’ (named by the experiment_name parameter) saved to folder specified by output_path. In the Colab Notebook demo, this would be /content/DemoOutput/demo_experiment/.
Opening the above experiment folder you will find the following folder/file hierarchy:
DatasetComparisons- all statistical significance results and plots for comparing modeling performance across multiple ‘target datasets’ rundataCompBoxplots- all data comparison boxplots
hcc_data- all output specific to the first ‘target dataset’ analyzedCVDatasets- copies of all training and testing datasets in .csv format (as well as intermediate files ifoverwrite_cv=False)exploratory- all phase 1 exploratory data analysis (EDA) output, files at this level are post-processed EDA outputinitial- all pre-processed EDA outputunivariate_analyses- all univariate analysis results and plots
feature_selection- all phase 3 & 4 output (feature importance estimation and feature selection)multisurf- MultiSURF scores and a summary figuremutual_information- mutual information scores and a summary figure
model_evaluation- all model evaluation output (phase 6)feature_importance- all model feature importance estimation scores and figuresmetricBoxplots- all evaluation metric boxplots comparing algorithm performancepickled_metrics- all evaluation metrics pickled separately for each algorithm and CV dataset combostatistical_comparisons- all statistical significance results comparing algorithm performance
models- all model output (phase 5), including pickled model objects and selected hyperparameter settings for each algorithm and CV dataset combopickledModels- all models saved as pickled objects
scale_impute- all trained imputation and scaling maps saved as pickled objects
hcc_data_custom- contains all output specific to the second ‘target dataset’ analyzedHas the same folder hierarchy as
hcc_dataabove with the addition of areplicationfolderreplication- all phase 8 (i.e. replication) output for the second ‘target dataset’ analyzedhcc_data_custom_rep- all replication output for the this specific ‘replication dataset’ (in this demo there was only one)exploratory- all exploratory data analysis (EDA) output for this ‘replication dataset’, files at this level are post-processed EDA outputinitial- all pre-processed EDA output for this ‘replication dataset’
model_evaluation- all model evaluation output for this ‘replication dataset’metricBoxplots- all evaluation metric boxplots comparing algorithm performance for this ‘replication dataset’pickled_metrics- all evaluation metrics pickled separately for each algorithm and CV dataset combo (for this ‘replication dataset’)statistical_comparisons- all statistical significance results comparing algorithm performance (for this ‘replication dataset’)
jobs- contains cluster job submission files (empty if output ‘cleaning’ applied)jobsCompleted- contains cluster checks for job completion (empty if output ‘cleaning’ applied)logs- contains cluster job output and error logs (empty if output ‘cleaning’ applied)
Notice that the folders hcc_data and hcc_data_custom have similar contents, but represent the analysis for each ‘target’ dataset run at once with STREAMLINE. If a user were to include 4 datasets in the folder specified by the dataset_path parameter (each conforming to the Input Data Requirements) they would find 4 respective folders in their experiment fold, each named after a respective dataset.
Output File Details
This section will take a deeper dive into the individual output files within an experiment folder.
PDF Report(s)
Testing Evaluation Report
When you first open the experiment folder, you will find the file demo_experiment_ML_Pipeline_Report.pdf. This is an automatically formatted PDF summarizing key findings during the model training and evaluation. It conveniently documents all STREAMLINE run parameters, and summarizes key results for data processing, processed data EDA, model evaluation, feature importance, algorithm comparisons, dataset comparisons, and runtime.
Replication Evaluation Report
A simpler ‘replication report’ is generated for each ‘replication dataset’ applied to the models trained by a single ‘target dataset’. You can find the demo replication report at the following path: /demo_experiment/hcc_data_custom/replication/hcc_data_custom_rep/demo_experiment_ML_Pipeline_Replication_Report.pdf. This report differs from the testing evaluation report in that it excludes the following irrelevant elements: (1) univariate analysis summary, (2) feature importance summary, (3) dataset comparison summary, and (4) runtime summary.
Experiment Meta Info
When you first open the experiment folder, you will also find algInfo.pickle and metadata.pickle which are used internally by STREAMLINE across most phases, as well as by the ‘Useful Notebooks’, covered in Doing More with STREAMLINE.
DatasetComparisons
At the beginning of the testing evaluation report, each dataset is assigned an abbreviated designation of ‘D#’ (e.g. D1, D2, etc) based on the alphabetical order of each dataset name. These designations are used in some of the files included within this folder.
Statistical Significance Comparisons
When you first open this folder you will find .csv files containing all statistical significance results comparing modeling performance across two or more ‘target datasets’ run at once with STREAMLINE.
STREAMLINE applies three non-parametric tests of significance:
Kruskal Wallis one-way analysis of variance - used for comparing two or more independent samples of equal or different sample sizes
Mann-Whitney U test (aka Wicoxon rank-sum test) - used for pair-wise independent sample comparisons
Wilcoxon signed-rank test - used for pair-wise dependent sample comparisons.
The .csv files in this folder include the above significance tests’ results comparing model performance between ‘target datasets’:
BestComparefiles: (1) apply the given test to each evalution metric, (2) only compares the models from the ‘top-performing-algorithm’ for a given metric/dataset - determined by which had the best median metric value for in a ‘sample’ (3) where a ‘sample’ is the set of k trained CV models for a given algorithmKruskalWallisfiles: (1) applies Kruskal Wallis to each evalution metric for a given algorithm across dataset ‘samples’, (2) where a ‘sample’ is the set of k trained CV models for that algorithmMannWhitneyfiles: (1) applies Mann-Whitney to each evalution metric for a given algorithm examining pairs of dataset ‘samples’, (2) where a ‘sample’ is the set of k trained CV models for that algorithmWilcoxonRankfiles: (1) applies Wilcoxon to each evalution metric for a given algorithm examining pairs of dataset ‘samples’, (2) where a ‘sample’ is the set of k trained CV models for that algorithm
dataCompBoxplots
This folder contains two different types of box plots comparing dataset performance:
DataCompare: (1) one plot for each combination of algorithm + evaluation metric (only ROC-AUC, and PRC-AUC metrics), (2) the ‘sample’ making up each individual box-and-whisker is the set of k trained CV models for that algorithmDataCompareAllModels: (1) one plot for each evaluation metric (all 16 classification metrics), (2) the ‘sample’ making up each individual box-and-whisker is the set of median algorithm performances, (3) lines are overlaid on the boxplot to illustrate differences in median performance between datasets for all algorithms.
hcc-data_custom
We will focus on the hcc_data_custom folder to walk through the remaining files, since (unlike the hcc_data folder) also includes the results of a replication analyis. However, note that you will find mostly the same set of files within hcc_data or for any uniquely named dataset in the folder specified by the dataset_path parameter.
The only file you will see when opening this folder is runtimes.csv which documents STREAMLINE’s runtime on different phases and machine learning modeling algorithms.
CVDatasets
This folder contains all training and testing datsets (named as [DATANAME]_CV_[PARTITION]_[Train or Test].csv). These cross validation (CV) datasets have undergone processing, imputation, scaling, and feature selection, and are the datasets used for model training and evaluation in phase 5.
Additionally, if overwrite_cv and del_old_cv were both False, you will see two additional sets of CV datasets with either CVOnly or CVPre in their filenames. These are intermediary versions of the CV datasets (included as a further sanity check), allowing users to examine how these datasets have changed prior to phase 2 (scaling and imputation), and phase 4 (feature selection).CVOnly identifies CV datasets that have undergone phase 1 processing (i.e. cleaning, feature engineering, and CV partitioning). CVPre identifies CV datasets that have additionally undergone phase 2 (scaling and imputation).
exploratory
We will begin by explaining the files you see when first opening this folder. All of these files represent exploratory data analysis (EDA) of the ‘processed data’ (i.e. after automated cleaning and feature engineering).
exploratory (plots)
ClassCountsBarPlot- a simple bar plot illustrating class balance or imbalanceDataMissingnessHistogram- a histogram illustrating the frequency of data missingness across featuresFeatureCorrelations- a pearson feature correlation heatmap
exploratory (.csv)
ClassCounts- documents the number of instances in the data for each classcorrelation_feature_cleaning- documents feature pairs that met thecorrelation_removal_thresholdidentifying which feature was retained vs deleted from the datasetDataCounts- documents dataset counts for number of instances, features, feature types, and missing valuesDataMissingness- documents missing value counts for all columns in the datasetDataProcessSummary- documents incremental changes to instance, feature, feature type, missing value, and class counts during the individual cleaning and feature engineering steps in phase 1DescribeDataset- output from standard pandasdescribe()functionDtypesDataset- output from standard pandasdtypes()functionFeatureCorrelations- documents all pearson feature correlationsMissingness_Engineered_Features- documents any newly engineered ‘missingness’ features added to the dataset based on thefeatureeng_missingnesscutoffMissingness_Feature_Cleaning- documents any features that have been removed from the data because their missingness was >=cleaning_missingnessNumerical_Encoding_Map- documents the numerical encoding mapping for any binary text-valued features in the datasetNumUniqueDataset- output from standard pandasnunique()functionOriginalFeatureNames- documents all original feature names from the ‘target dataset’ prior to any processingprocessed_categorical_features- documents all processed feature names that were be treated as categoricalprocessed_quantitative_features- documents all processed feature names that were be treated as quantitativeProcessedFeatureNames- documents all feature names for the processed ‘target dataset’
exploratory (pickle)
A variety of other pickle files can be found in this folder, used internally for data processing in the replication phase.
initial
This subfolder includes a subset of the same files found in exploratory, however these files represent the ‘initial’ exploratory data analysis (EDA) prior to cleaning and feature engineering.
univariate analysis
This subfolder includes plots and a .csv report focused on exploratory univariate analyses on the given processed ‘target dataset’.
univariate analysis (plots)
Barplotplots: simple barplots illustrating the relationship between a given categorical feature and outcome if the Chi Square Test was significant based onsig_cutoffBoxplotplots: simple boxplots illustrating the relationship between a given quantitative feature and outcome if the Mann-Whitney U test was significant based onsig_cutoff
univariate analysis (.csv)
Univariate_Signifiance.csv documents the p-value, test statistic, and test name applied across all processed features in the ‘target dataset’
feature_selection
Includes all output for phases 3 and 4 (feature importance estimation and feature selection).
When first opening this folder you will find
InformativeFeatureSummary.csvwhich summarizes feature counts kept or removed during feature selection (i.e. Informative vs. Uninformative) for each individual CV partition.
multisurf
This subfolder includes (1) .csv files with MultiSURF scores for each CV partition and (2) TopAverageScores, a plot of the top (top_fi_features) features (based on median MultiSURF score over CV paritions)
mutual_information
This subfolder includes (1) .csv files with mutual information scores for each CV partition and (2) TopAverageScores, a plot of the top (top_fi_features) features (based on median mutual information score over CV paritions)
model_evaluation
We will begin by explaining the files you see when first opening this folder.
model_evaluation (plots)
[ALGORITHM]_ROCplots: reciever operating characteristic (ROC) plot for a given algorithm illustrating performance across all CV partitions (i.e. ‘folds’), as well as the mean ROC curve and +- 1 standard deviation of the CV partitions.[ALGORITHM]_PRCplots: precision-recall curve (ROC) plot for a given algorithm illustrating performance across all CV partitions (i.e. ‘folds’), as well as the mean PRC and +- 1 standard deviation of the CV partitions.Summary_ROCplot - reciever operating characteristic (ROC) plot comparing mean (CV partition) ROC curves across all algorithms.Summary_PRCplot - precision-recall curve (ROC) plot comparing mean (CV partition) PRCs across all algorithms.
model_evaluation (.csv)
[ALGORITHM]_performance: documents the 16 model performance metrics for each CV partition for this algorithmSummary_performance_mean- documents the 16 performance metrics (mean across CV partition) for each algorithmSummary_performance_median- documents the 16 performance metrics (median across CV partition) for each algorithmSummary_performance_std- documents the 16 performance metrics (standard deviation across CV partitions) for each algorithm
feature_importance
This subfolder includes all model specific feature importance outputs including plots and .csv files for the ‘target dataset’.
Plots are as follows:
Compare_FI_Norm- composite feature importance plot illustrating normalized model feature importance scores across all algorithms run.top_fi_featuresfeatures are displayed and ranked based on the across-algorithm sum of mean normalized and weighted feature importance scores. This weighting is based on the model performance indicated by ‘metric_weight’.Compare_FI_Norm_Weight- composite feature importance plot illustrating normalized and weighted model feature importance scores across all algorithms run.top_fi_featuresfeatures are displayed and ranked based on the across-algorithm sum of mean normalized and weighted feature importance scores. This weighting is based on the model performance indicated by ‘metric_weight’.[ALGORITHM]_boxplotplots: boxplot of model feature importance scores (for a given algorithm).top_fi_featuresfeatures are displayed, ranked by mean model feature importance scores (across CV partitions).[ALGORITHM]_histogramplots: histogram illustrating the distribution of the mean (across CV partitions) model feature importance scores (for a given algorithm).
The .csv files are as follows:
[ALGORITHM]_FI: documents all model feature importance estimates (for the full list of processed features) for each CV partition (for a given algorithm)
metricBoxplots
This subfolder contains separate boxplox plots for each model evaluation metric. Each plot compares the set of algorithms run across all CV partitions.
pickled_metrics
This subfolder contains pickle files (used internally) to store evaluation metrics for each algorithm and CV partition combination.
statistical_comparisons
This subfolder contains .csv files documenting statistical significance tests comparing algorithm performance on the given ‘target dataset’.
KruskalWallis- documents Kruskal Wallis applied to each evalution metric between algorithm ‘samples’, (2) where a ‘sample’ is the set of k trained CV models for that algorithmMannWhitneyUfiles: documents Mann-Whitney applied to a given evalution metric between pairs of algorithm ‘samples’, (2) where a ‘sample’ is the set of k trained CV models for that algorithmWilcoxonRankfiles: documents Wilcoxon applied to a given evalution metric between pairs of algorithm ‘samples’, (2) where a ‘sample’ is the set of k trained CV models for that algorithmNote:
MannWhitneyUandWilcoxonRankfiles are only generated for a given evaluation metric if theKruskalWallistest for that metric was significant.
models
Upon opening this folder you will see .csv files for each algorithm and CV partition combination documenting the ‘best’ hyperparameter settings identified by Optuna and used to train each respective final model.
Also included is the pickledModels subfolder containing all trained and pickled model objects for each algorithm and CV partition combination. Beyond the testing and replication data performance evaluation output by STREAMLINE, these models can be unpickled and applied in the future to document (1) training performance on the training datasets, (2) further replication datasets, and (3) applied to unlabled data to make outcome predictions.
replication
This folder will include a subfolder for every ‘replication dataset’ being applied to a given ‘target dataset’. In the demo, this only includes hcc_data_custom_rep. Within this folder you will find a subset of relevant the folders and output files we have already covered; plotting and documenting the given replication dataset and the findings when evaluating all trained models using this replication dataset. This includes the PDF Replication Evaluation Report and a ‘processed’ copy of the given ‘replication dataset’.
runtime
This folder includes .txt files documenting the runtimes spent on different phases and algorithms within STREAMLINE. As previously mentioned, these times are summarized within runtimes.csv generated for each ‘target dataset’ (e.g. /demo_experiment/hcc_data_custom/runtimes.csv).
scale_impute
This folder includes all pickled trained mappings used internally for missing value imputation and applying standard scaling to new data.
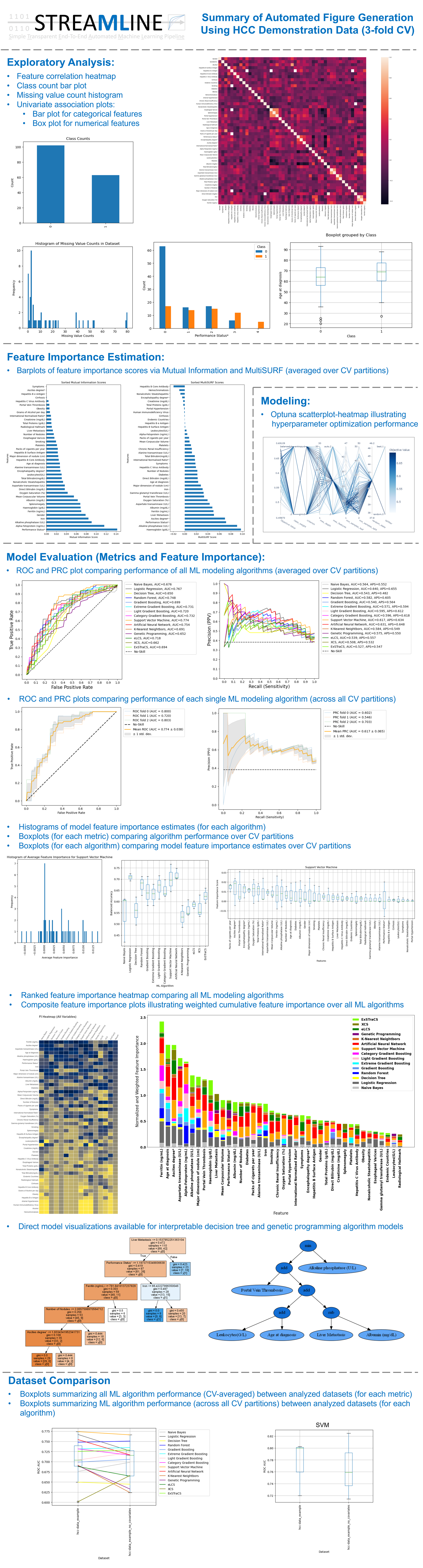
Figures Summary
Below is an example overview of the different figures generated by STREAMLINE for binary classification data. Note that these current images were generated from the Beta 0.2.5 release, and have been improved, updated and expanded since.